Computational Tools
Bacteria Segmentation Software:
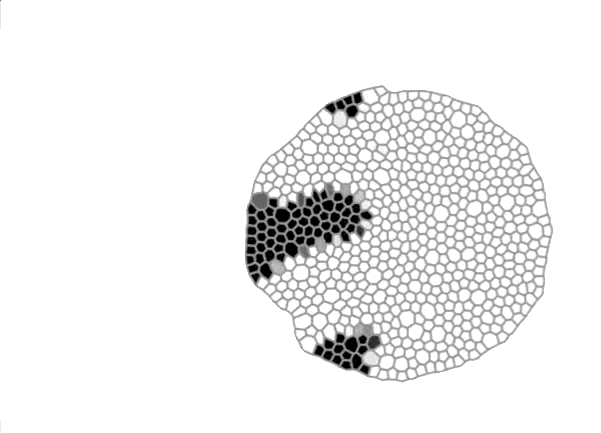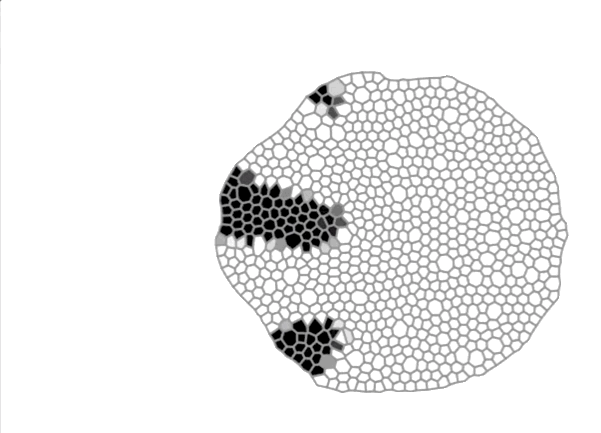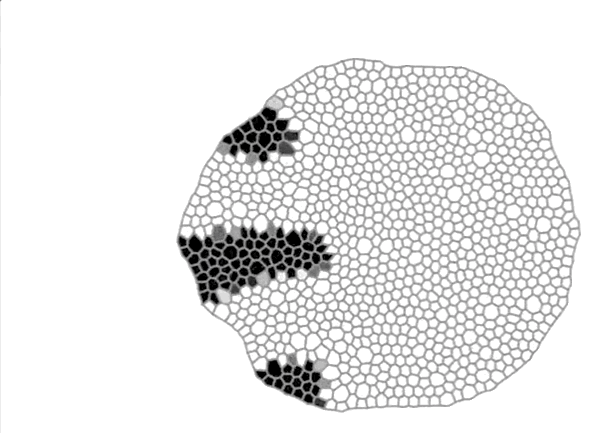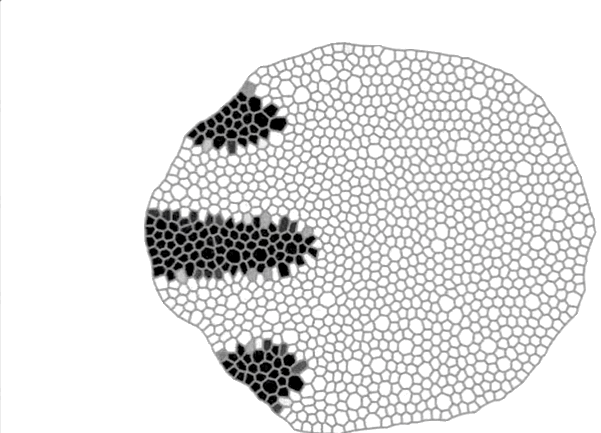
This tool performs machine-learning-based segmentation and lineage tracking of dense bacterial population using FiJi (ImageJ) as a front-end. This image shows how reliable is the software.
More information can be found in this publication or at the webpage of the project, including example images and experimental protocols (click on the icon)
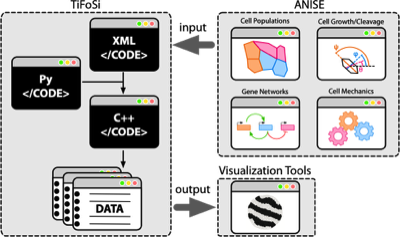
TiFoSi:
TiFoSi (Tissues: Forces and Signaling) simulates the dynamics of epithelial tissues in 2D. TiFoSi includes signaling (e.g. protein expression) and mechanical modules (and feedbacks between them if needed), and allows for different cell populations, configurable cell cycle and cleavage properties, and many more functionalities. TiFoSi uses configuration files that are converted into efficient C++ code.
Further information can be found in this publication or at the webpage of the project (click on the icon)
ANISE:
ANISE (grAphical coNfigurator of TIFOSI In Silico Experiments) is a web-app to easily create/edit the XML configuration files of TIFOSI. ANISE also includes a visual design tool to model gene regulatory interactions. The app can be installed/run locally (any OS) or deployed to a server. As an example of the later, this server is available for users.
Further information can be found in this publication or at the webpage of the TIFOSI project (click on the icon)

Computational solutions for Systems Biology
Part of the activities of our group are related with developing computational solutions in the field of Systems Biology. Thus, we have developed a tool to segment and track bacterial populations growing into crowded conditions. This tool leverages the machine learning capabilities of FiJi (ImageJ) provided through Weka and custom-made Python scripts. On the other hand, we have developed a tool to implement simulations of tissues using a vertex model approach. This software allows for mechano-signalling feedbacks and multiple cell populations (on top of other functionalities) and it is computationally efficient (fast!).
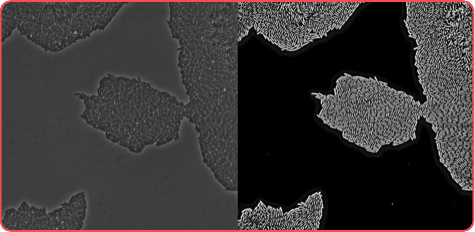
Phase contrast image
Segmented image
Buceta’s Lab
